
A step-by-step derivation of the Poisson log-likelihood and maximum likelihood estimators for gene expression counts in single-cell data.
Nov 17, 2025
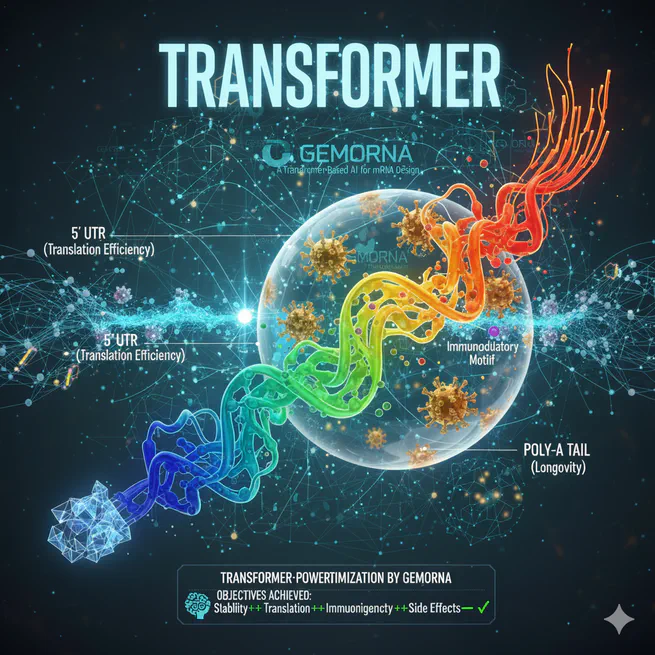
Exploring the architecture, training strategies, and surprising emergent capabilities of GEMORNA models for comprehensive mRNA design.
Aug 31, 2025
A comprehensive technical guide to edgeR's statistical framework, covering dispersion estimation, differential expression testing, and quasi-likelihood F-tests with detailed numeric examples.
May 27, 2025
Exploring how scDesign3 uses Gaussian and vine copulas to model complex dependencies between genes in single-cell RNA sequencing data.
Mar 22, 2025
Here continues to dive into the mathematics crucial for effective GSEA analysis and machine learning applications in bioinformatics.
Jan 15, 2024
Here continues to dive into the mathematics crucial for effective GSEA analysis
Jan 15, 2024
Learn why using all genes with proper ranking statistics is crucial for GSEA analysis, and get ready-to-use R code for different differential expression tools.
Jan 15, 2024